With Celline, NanoTag introduced a unique proprietary technology for the time-efficient discovery of single-domain antibodies.
In a single affinity selection step, a solid support displaying the target protein is used to purify target-specific B lymphocytes isolated from a pool of PBMCs. After washing, individual B-cells presenting target-specific antibodies on their surface are selectively eluted and further processed. The genetic information containing the sequence for the displayed antibodies is extracted, amplified by RT-PCR with sdAb-specific primers, and cloned into a prokaryotic expression vector. Identified sdAb clones are finally analyzed by ELISA or target-specific functional assays. Importantly, the washing stringency and elution mode during the initial affinity selection step can be customized within a wide range of conditions. This allows for the enrichment of B cells presenting antibodies with desired properties already in this stage.
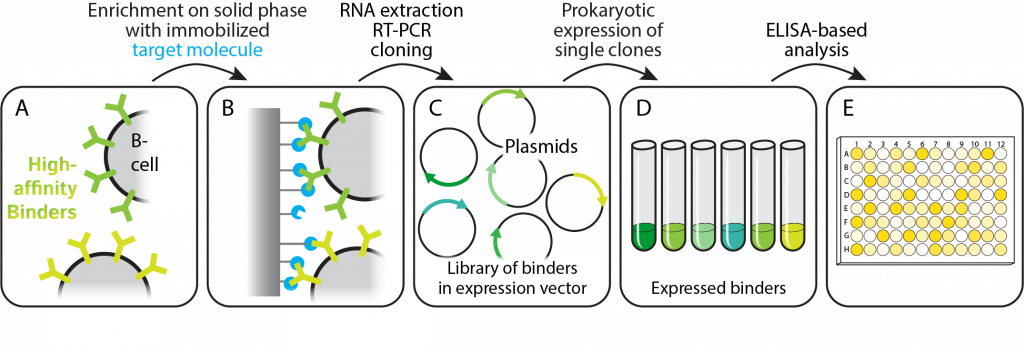
In terms of topology, Celline is an ideal camelid sdAb discovery platform when downstream assays and technologies require sdAbs to recognize proteins presented on the surface of target cells, e.g., surface-protein directed purification of blood cells or CAR-T technologies. Celline may therefore be advantageous especially for the selection of sdAbs intended for such applications.
Celline is a single-step method and ideally suited for the time-efficient development of sdAbs. After immunization, single binders can be identified within 5-8 working days.
NanoTag is currently implementing additional steps rendering the selection process specific to camelid B cells presenting camelid heavy-chain antibodies (IgG2/3 isotypes), which are the precursors of single-domain antibodies.